Water collapse (implicit)
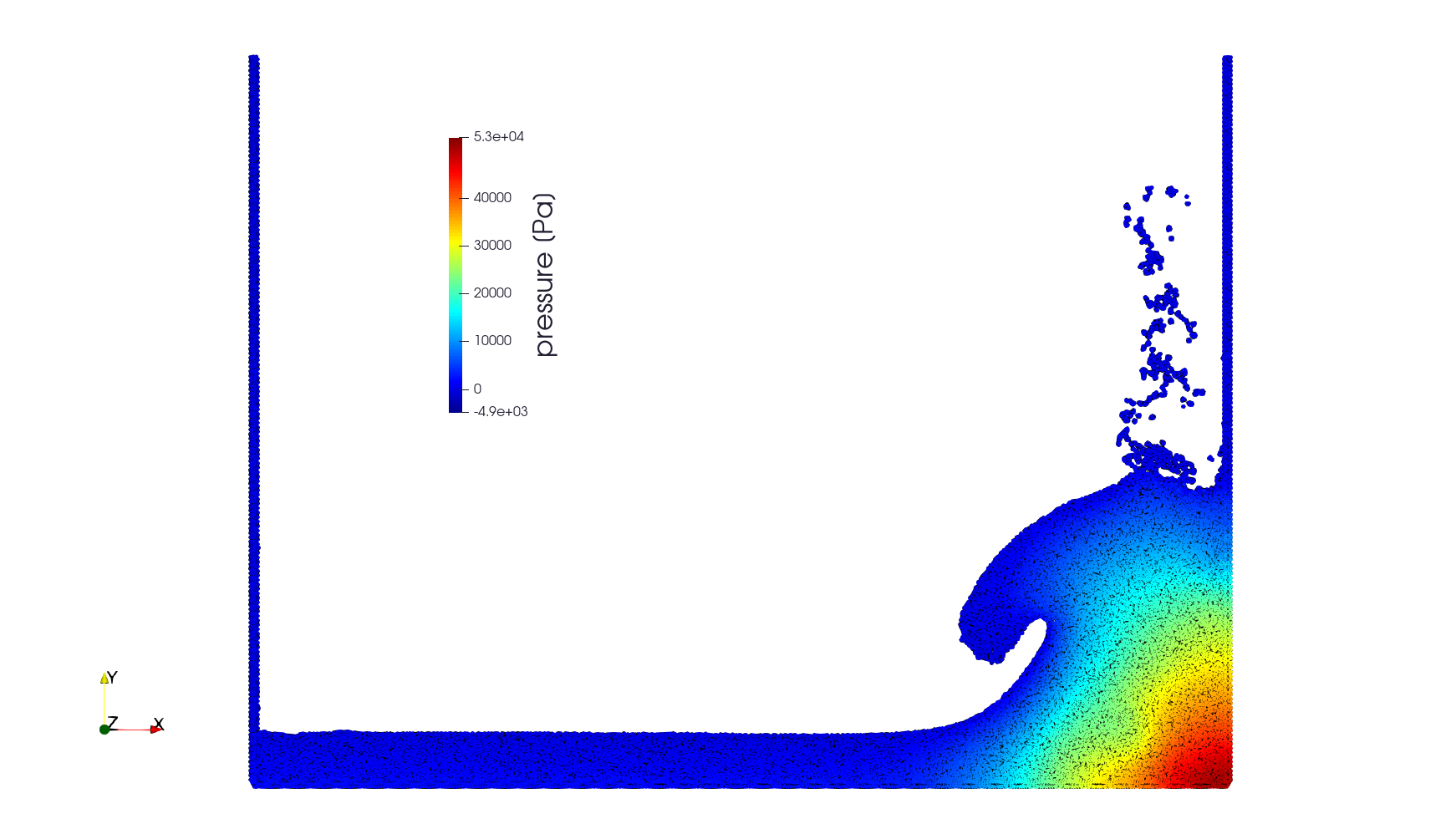
Simulation of a water column, this time using incompressible SPH. This works by first applying all non-pressure forces to find intermeidate velocity $\mathbf{v}^*$. The pressure-corrected velocity
$\mathbf{v} = \mathbf{v}^* - \frac{\delta t}{\varrho} \nabla P$
is found such that $\mathbf{v}$ is a divergence-free vector field. Taking divergence of both sides leads to a poisson problem
$- \Delta P = -\frac{\varrho}{\delta t} \nabla \cdot \mathbf{v}^*.$
A homogeneous Dirichlet condition for pressure is prescribed at the boundary. Free surface can be identified using a trick invented by Lee (2008). A parameter is defined
$\lambda = 1 - \frac{1}{d} \nabla \cdot {\mathbf{r}}$
which is zero inside the fluid but positive on the boundary. Boundary condition can be implemented smoothly by adding a penalty term:
$- \Delta P + C_\mathrm{free} \lambda P = -\frac{\varrho}{\delta t} \nabla \cdot \mathbf{v}^*.$
To get a particle approximation, we replace $\Delta$ with a Morris operator and $\nabla \cdot$ with the usual divergence operator. This yields a system with a positive definite matrix.
module collapse_dry_implicit
using Printf
using SmoothedParticles
using CSV
using DataFrames
using LinearAlgebra
using IterativeSolvers
using IncompleteLU
using Parameters
using PlotsConstant parameters
##kernel functions
const kernel = spline23
const Dkernel = Dspline23
const rDkernel = rDspline23
const dim = 2
const dr = 1.0e-2 # average particle distance (decrease to refine, increase to speed up)
const h = 2.8*dr # size of kernel support
const rho = 1000.0 # fluid density
const g = -9.8*VECY # gravitational acceleration
const mu = 8.4e-4 # dynamic viscosity
const m = dr^dim*rho # particle mass
const C_free = 10.0 # free surface penalty coefficient
const v_char = 5.0 # char velocity
##geometry parameters
const water_column_width = 1.0
const water_column_height = 2.0
const box_height = 3.0
const box_width = 4.0
const nlayers = 3.5
const wall_width = nlayers*dr
##temporal parameters
const dt = 0.1*h/v_char
const t_end = 2.0
const dt_frame = max(dt, t_end/200)
##labels for particle types
const FLUID = 0.
const WALL = 1.
const DUMMY = 2.Particle variables
@with_kw mutable struct Particle <: AbstractParticle
x::RealVector = VEC0 #position
v::RealVector = VEC0 #velocity
Dv::RealVector = VEC0 #acceleration
P::Float64 = 0.0 #pressure
div::Float64 = 0.0 #divergence of velocity
L::Float64 = 0.0 #diagonal element of projection matrix
lambda::Float64 = 0.0 #free surface indetifier
type::Float64 #particle type
endGeometry
function make_system()
grid = Grid(dr, :hexagonal)
box = Rectangle(0., 0., box_width, box_height)
fluid = Rectangle(0., 0., water_column_width, water_column_height)
walls = Specification(BoundaryLayer(box, grid, 1.2*dr), x -> (x[2] < box_height))
dummy = Specification(BoundaryLayer(box, grid, nlayers*dr) - walls, x -> (x[2] < box_height))
sys = ParticleSystem(Particle, fluid + dummy + walls, h)
generate_particles!(sys, grid, fluid, x -> Particle(x=x, type=FLUID))
generate_particles!(sys, grid, walls, x -> Particle(x=x, type=WALL))
generate_particles!(sys, grid, dummy, x -> Particle(x=x, type=DUMMY))
create_cell_list!(sys)
return sys
endParticle interactions
function initialize!(p::Particle)
if p.type == FLUID
p.x += dt*p.v
p.v += dt*g
end
p.div = 0.0
p.L = 0.0
p.lambda = 1.0
end
function viscous_force!(p::Particle, q::Particle, r::Float64)
p.Dv += 2.0*m*mu*rDkernel(h,r)/rho^2*(p.v - q.v)
end
function internal_force!(p::Particle, q::Particle, r::Float64)
p.Dv -= m*rDkernel(h,r)*(p.P + q.P)/rho^2*(p.x - q.x)
end
function accelerate!(p::Particle)
if p.type == FLUID
p.v += dt*p.Dv
end
p.Dv = VEC0
endFunctions to build the linear system
function div_L_lambda!(p::Particle, q::Particle, r::Float64)
rDk = rDkernel(h,r)
p.div += -SmoothedParticles.dot(p.x - q.x, p.v - q.v)*m*rDk
p.L += -2.0*m/rho*rDk
p.lambda += m/rho*rDk*r^2/dim
end
function projection_matrix(p::Particle, q::Particle, r::Float64)::Float64
if p == q
if p.type == FLUID
return h^2*p.L + C_free*max(p.lambda, 0.)
else
return h^2*p.L
end
end
return 2.0*h^2*m/rho*rDkernel(h,r)
end
function projection_vector(p::Particle)::Float64
return -h^2*p.div/dt
endExtract global variables
Variables of interest are total energy, water column height and wavefront location.
function energy(p::Particle)::Float64
kinetic = 0.5*m*SmoothedParticles.dot(p.v, p.v)
potential = -m*SmoothedParticles.dot(g, p.x)
return kinetic + potential
end
function get_globals(sys::ParticleSystem)::NTuple{3,Float64}
H = 0.0 # height of water column
X = 0.0 # wavefront x-coordinate
E = 0.0 # total energy
for p in sys.particles
if p.type == FLUID
X = max(X, p.x[1]/water_column_width)
end
if p.type == FLUID && 2.0 > p.x[1] > h
H = max(H, p.x[2]/water_column_height)
end
E += energy(p)
end
return (X,H,E)
endTime iteration
function main()
sys = make_system()
out = new_pvd_file("results/collapse_dry_implicit")
P = ParticleField(sys, :P)
ts = []
Xs = []
Hs = []
@time for k = 0 : Int64(round(t_end/dt))
if (k % Int64(round(dt_frame/dt)) == 0)
@printf("t = %.6e s ", k*dt)
println("(",round(100*k*dt/t_end),"% complete)")
(X, H, E) = get_globals(sys)
@printf("energy = %.6e J\n", E)
@printf("\n")
push!(Xs, X)
push!(Hs, H)
push!(ts, k*dt*sqrt(-2*g[2]))
save_frame!(out, sys, :v, :P, :type)
end
apply!(sys, initialize!)
create_cell_list!(sys)
apply!(sys, viscous_force!)
##assemble linear system and solve for pressure
apply!(sys, div_L_lambda!)
A = assemble_matrix(sys, projection_matrix)
b = assemble_vector(sys, projection_vector)
try
#P .= A\b
P .= cg(A, b)#; Pl = ilu(A; τ = 0.1))
catch
save_pvd_file(out)
error("Unable to solve linear system.")
end
apply!(sys, internal_force!)
apply!(sys, accelerate!)
end
save_pvd_file(out)
data = DataFrame(time = ts, X = Xs, H = Hs)
CSV.write("results/collapse_dry_implicit/data.csv", data)
@info "drawing a plot with results"
make_plot()
endCompare computed results to the book by Violeau.
function make_plot()
data = CSV.read("results/collapse_dry_implicit/data.csv", DataFrame)
X_VIO = CSV.read("reference/dambreak_X_Violeau.csv", DataFrame)
X_KOS = CSV.read("reference/dambreak_X_Koshizuka.csv", DataFrame)
H_VIO = CSV.read("reference/dambreak_H_Violeau.csv", DataFrame)
H_KOS = CSV.read("reference/dambreak_H_Koshizuka.csv", DataFrame)
p1 = plot(data.time, data.X, label = "SmoothedParticles.jl", xlims = (0., 3.0))
scatter!(p1, X_VIO.time, X_VIO.X, label = "Violeau")
scatter!(p1, X_KOS.time, X_KOS.X, label = "Koshizuka&Oda", markershape = :square)
savefig(p1, "results/collapse_dry_implicit/dambreak_X.pdf")
p2 = plot(data.time, data.H, label = "SmoothedParticles.jl", xlims = (0., 3.0))
scatter!(p2, H_VIO.time, H_VIO.H, label = "Violeau")
scatter!(p2, H_KOS.time, H_KOS.H, label = "Koshizuka&Oda", markershape = :square)
savefig(p2, "results/collapse_dry_implicit/dambreak_H.pdf")
end
endThis page was generated using Literate.jl.